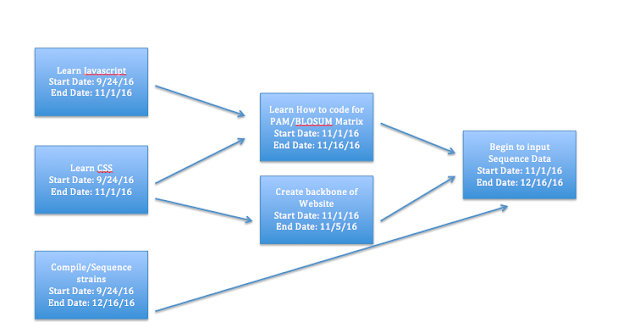
Updated Gantt Chart

Over the next three months I will be getting all the preliminary steps of creating a bioinformatics website out of the way. I will learn how to code in Javascript and CSS and then begin gathering the data that the website will organize. Finally, I will start the laying the foundations of the website.
Initial Project Planning:
Identify the Problem:
Lyme disease is constantly evolving and new strains are emerging. This makes it increasingly hard to treat because the strains are evolving to evade antibiotics. We need to sequence all existing strains and find which sections are increasing virulence and allowing the strains to evade the immune system and which sections are conserved.
Prior Art Survey:
There is a bioinformatics site which has compiled the sequences of 35 genomes of 8 LB species and 7 RF species. It is a manually curated site which includes comparisons of phylogeny, synteny and sequence alignments of orthologous sequences and intergenic spacers. (http://borreliabase.org/)
Literature Review:
Can be found in previous posts.
Solution Space Exploration:
Since a site already exists with with MSA, phylogeny and synteny. They do not have a complete genome for most strains. They also do not have the functions of each gene.
High Level Design:
I need to get in contact with the people who wrote the paper I read over the summer about lime disease before I start my project.
Tools:
If I plan to work off of the existing database for lyme disease I will need just a computer and possibly access to the NCBI database, in order to read the articles on lyme disease strains. I could access these through the library.
If I plan to work with sequencing that have not been sequenced yet, then I will need access to a lab or some sort of collaboration with a lab to sequence the rest of the genome.
I will need a mentor who knows about the lyme disease genome
Budget:
Most likely 0$
http://www.novusbio.com/diseases/lyme-disease
http://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471-2105-15-233
Bioinformatics Slide:
https://docs.google.com/presentation/d/17VzdZdIEADFSacXulYF5ocCSfWaYhgboPMJYiH3iaGA/edit?usp=sharing





